Publications

Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation
Mathieu Botte, Dongchun Ni, Stephan Schenck, Iwan Zimmermann, Mohamed Chami, Nicolas Bocquet, Pascal Egloff, Denis Bucher, Matilde Trabuco, Robert K. Y. Cheng, Janine D. Brunner, Markus A. Seeger, Henning Stahlberg & Michael Hennig@. Nature Communications 13, 1826 (2022). https://doi.org/10.1038/s41467-022-29459-2.
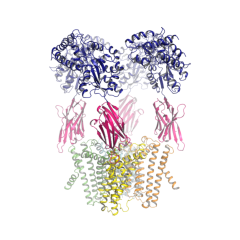
Structural basis for ion selectivity in TMEM175 K+ channels
Brunner JD@, Jakob RP, Schulze T, Neldner Y, Moroni A, Thiel G, Maier T, Schenck S@. Elife. 2020 Apr 8;9:e53683. doi: 10.7554/eLife.53683. PMID: 32267231; PMCID: PMC7176437.

Production and Application of Nanobodies for Membrane Protein Structural Biology.
Brunner JD@, Schenck S. Methods Mol Biol. 2020;2127:167-184. doi: 10.1007/978-1-0716-0373-4_12. PMID: 32112322.

Preparation of Proteoliposomes with Purified TMEM16 Protein for Accurate Measures of Lipid Scramblase Activity
Brunner JD@, Schenck S. Methods Mol Biol. 2019;1949:181-199. doi: 10.1007/978-1-4939-9136-5_14. PMID: 30790257.

Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Paulino C, Neldner Y, Lam AK, Kalienkova V, Brunner JD, Schenck S, Dutzler R@. Elife. 2017 May 31;6:e26232. doi: 10.7554/eLife.26232. PMID: 28561733; PMCID: PMC5470873.

Structural basis for phospholipid scrambling in the TMEM16 family
Brunner JD, Schenck S, Dutzler R@. Curr Opin Struct Biol. 2016 Aug;39:61-70. doi: 10.1016/j.sbi.2016.05.020. Epub 2016 Jun 10. PMID: 27295354.

X-ray structure of a calcium-activated TMEM16 lipid scramblase
Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R@. Nature. 2014 Dec 11;516(7530):207-12. doi: 10.1038/nature13984. Epub 2014 Nov 12. PMID: 25383531.
Calcium-activated proteins visualized - News and views by Matt Whorton
